- About the Lab
- Local Hardware Platform
- Remote Hardware Platform
- Software Platform
Pan-omics
"Panomics" or 'pan-omics' is a term used to refer to the range of molecular biology technologies including genomics, proteomics, metabolomics, transcriptomics, and so forth, or the integration of their combined use.
Genomics study the genomes of organisms. Cognitive genomics examines the changes in cognitive processes associated with genetic profiles.
Transcriptomics study of transcriptomes, their structures and functions. Transcriptome is the set of all RNA molecules, including mRNA, rRNA, tRNA, and other non-coding RNA, produced in one or a population of cells.
Proteomics is the large-scale study of proteins, particularly their structures and functions. Proteome is the entire complement of proteins, including the modifications made to a particular set of proteins, produced by an organism or system.
Metabolomics is the scientific study of chemical processes involving metabolites. It is a "systematic study of the unique chemical fingerprints that specific cellular processes leave behind", the study of their small-molecule metabolite profiles.
Big Data
Our lab focus on solving the challenge of big data management and its application to human healthcare.
The information management big data and analytics capabilities include :
Data Management & Warehouse: Gain industry-leading database performance across multiple workloads while lowering administration, storage, development and server costs; Realize extreme speed with capabilities optimized for analytics workloads such as deep analytics, and benefit from workload-optimized systems that can be up and running in hours.
Hadoop System: Bring the power of Apache Hadoop to the enterprise with application accelerators, analytics, visualization, development tools, performance and security features.
Stream Computing: Efficiently deliver real-time analytic processing on constantly changing data in motion and enable descriptive and predictive analytics to support real-time decisions. Capture and analyze all data, all the time, just in time. With stream computing, store less, analyze more and make better decisions faster.
Content Management: Enable comprehensive content lifecycle and document management with cost-effective control of existing and new types of content with scale, security and stability.
Information Integration & Governance: Build confidence in big data with the ability to integrate, understand, manage and govern data appropriately across its lifecycle.
Dell Precision Tower 5810 Workstation
| |
| Intel® Xeon® processors: Power through the most demanding, interactive applications quickly with a new generation of single-socket performance with the Intel® Xeon® processor E5-1600 v3 series. The Dell Precision Tower 5810 delivers up to 75 percent better performance than previous generations for iterative design and prototyping.
|
The Milky Way II Supercomputer System

ØThe Milky Way II supercomputer system, developed by the National University of Defense Technology (NUDT), is the outstanding achievement of the National 863 Program.
ØWith 16,000 computer nodes, each comprising two Intel Ivy Bridge Xeon processors and three Xeon Phi coprocessor chips, it represents the world's largest installation of Ivy Bridge and Xeon Phi chips, counting a total of 3,120,000 cores. Each of the 16,000 nodes possess 88 gigabytes of memory (64 used by the Ivy Bridge processors, and 8 gigabytes for each of the Xeon Phi processors). The total CPU plus coprocessor memory is 1,375 TiB (approximately 1.34 PiB).
ØDuring the testing phase, the Milky Way II supercomputer system was laid out in a non-optimal confined space. When assembled at its final location, the system will have a theoretical peak performance of 54.9 petaflops. At peak power consumption, the system itself would draw 17.6 megawatts of power. Including external cooling, the system would draw an aggregate of 24 megawatts. The computer complex would occupy 720 square meters of space.
ØThe front-end system consists of 4096 Galaxy FT-1500 CPUs, a SPARC derivative designed and built by NUDT. Each FT-1500 has 16 cores and a 1.8 GHz clock frequency. The chip has a performance of 144 gigaflops and runs on 65 watts. The interconnect, called the TH Express-2, designed by NUDT, utilizes a fat tree topology with 13 switches each of 576 ports.
| Perl: Perl is an ideal web programming language due to its text manipulation capabilities and rapid development cycle. |
| R: R provides a wide variety of statistical (linear and nonlinear modelling, classical statistical tests, time-series analysis, classification, clustering, …) and graphical techniques, and is highly extensible. |
| IPA: IPA unlock the insights buried in experimental data by quickly identifying relationships, mechanisms, functions, and pathways of relevance. |
| PHP: PHP is a server scripting language, and a powerful tool for making dynamic and interactive Web pages. |
| Apache: The Apache HTTP Server is the world's most widely used web server software, which is the first web server software to serve more than 100 million websites. |
| MySQL: MySQL is one of the world's most widely used relational database management system (RDBMS). |
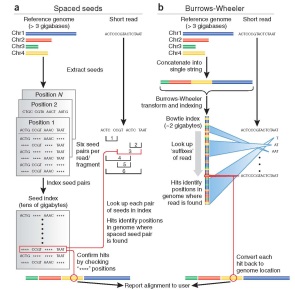
| BWA: BWA is a software package for mapping low-divergent sequences against a large reference genome, such as the human genome. |
| Bowtie: Bowtie is a fast short aligner using an algorithm based on the Burrows-Wheeler transform and the FM-index. |
| Cufflinks: Cufflinks is appropriate to measure global de novo transcript isoform expression. |
| Samtools: Samtools is a suite of programs for interacting with high-throughput sequencing data. |
| GATK: The Genome Analysis Toolkit or GATK is a software package developed at the Broad Institute to analyze high-throughput sequencing data. |
| SIFT: SIFT predicts whether an amino acid substitution affects protein function. |
| PolyPhen2: PolyPhen-2 is a tool which predicts possible impact of an amino acid substitution on the structure and function of a human protein using straightforward physical and comparative considerations. |
| PANTHER: The PANTHER Classification System was designed to classify proteins (and their genes) in order to facilitate high-throughput analysis. |